Prediction Model
In the first part of the project we make use of Compartmental models. Compartmental models simplify the mathematical modelling of infectious diseases. The population is assigned to compartments with labels - for example, S, I, or R, (Susceptible, Infectious, or Recovered). People may progress between compartments. The order of the labels usually shows the flow patterns between the compartments; for example SEIS means susceptible, exposed, infectious, then susceptible again.
These Models try to predict things such as how a disease spreads, or the total number infected, or the duration of an epidemic, and to estimate various epidemiological parameters such as the reproductive number. Such models can show how different public health interventions may affect the outcome of the epidemic, e.g., what the most efficient technique is for issuing a limited number of vaccines in a given population.
Over the time of this project, we plan to improve upon the SIR and SIER models in predicting the infected population and death rate.
The SIR Model
The SIR model is one of the simplest compartmental models, and many models are derivatives of this basic form. The model consists of three compartments:
- S: The number of susceptible individuals. When a susceptible and an infectious individual come into "infectious contact", the susceptible individual contracts the disease and transitions to the infectious compartment.
- I: The number of infectious individuals. These are individuals who have been infected and are capable of infecting susceptible individuals.
- R for the number of removed (and immune) or deceased individuals. These are individuals who have been infected and have either recovered from the disease and entered the removed compartment, or died. It is assumed that the number of deaths is negligible with respect to the total population. This compartment may also be called "recovered" or "resistant".

This model is reasonably predictive for infectious diseases that are transmitted from human to human, and where recovery confers lasting resistance, such as measles, mumps and rubella.
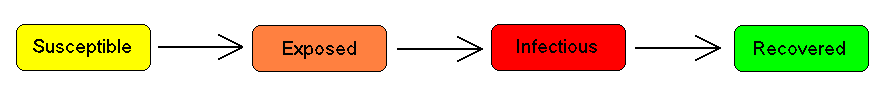
The SIER Model
For many important infections, there is a significant incubation period during which individuals have been infected but are not yet infectious themselves. During this period the individual is in compartment E (for exposed).
Source
Poisson Regression (GAM) Model
The goal is to provide the most accurate forecasts for COVID-19 in Canada for new cases, deaths and recovered cases. We'll be using an approach similar to that in the paper Spatiotemporal Dynamics, Nowcasting and Forecasting of COVID-19 in the United States which uses a generalized additive model with Poisson distribution using temporal variables (number of infected, susceptible cases at time 𝑡−1 ), spatial coordinates, time varying covariates (control measures), and constant covariates (demographic, economic, etc). For now we only use temporal variables 𝐼𝑖𝑡 and 𝑍𝑖𝑡.
Given variables
- 𝑌𝑖𝑡 : Current number of new case at time t for area i. Column "cases" in cases_timeseries_prov.csv
- 𝐼𝑖𝑡 : Cumulative number of active infectious cases. Column "active_cases" in active_timeseries_prov.csv
- 𝐶𝑖𝑡 : Cumulative confirmed cases up to time t. Column "cumulative_cases" in active_timeseries_prov.csv
- 𝑅𝑖𝑡 : Cumulative number of infections. Column "cumulative_recovered" in active_timeseries_prov.csv
Transformed variables
- 𝑁𝑖 : Population of region i. Column "population"
- Δ𝐷𝑖𝑡 : New deaths/recovered cases at time t. Column "removed"
- 𝑆𝑖𝑡 : Number of susceptible subjects at time t defined by 𝑆𝑖𝑡=𝑁𝑖−𝐶𝑖𝑡. Column "susceptible"
- 𝑍𝑖𝑡 : Log ratio of susceptible subjects 𝑍𝑖𝑡=𝑙𝑜𝑔(𝑆𝑖𝑡/𝑁𝑖). Column "percent_susceptible" without log transform.
We'll be applying a similar but simpler version of the poisson regression as proposed in the paper mentioned above. Currently it only accounts for temporal effects. For a given province 𝑖 we have the new cases modelled as:
and the new deaths/recovered modelled as: